Projects
Cryo-Electron Tomography
PI: Grant Jensen (Division of Biology and Biological Engineering)
SASE: Kyung Min Shin, Scholar
Cryo-electron microscopy (cryo-EM) technologies are revolutionizing structural biology, pushing the boundaries of biological targets that can be resolved across a range of length scales. Two modalities of cryo-EM – single particle reconstruction and micro-electron diffraction – have together enabled high-resolution structure determination of large complexes and small proteins, including many intractable to the widely-used method of X-ray crystallography. Electron cryotomography (ECT) is the third principal cryo-EM modality and has demonstrated great power to reveal the 3-D structures of viruses and macromolecular complexes in the native context of the cell. But vast potential remains untapped. This claim rests on three principles: (1) electrons offer key advantages over photons for imaging, (2) 3-D information is superior to 2-D, and (3) cryopreservation avoids artifacts that can distort native structure. ECT takes advantage of all three principles and will thus offer a powerful method for many cell and structural biologists once two technical limitations are overcome.
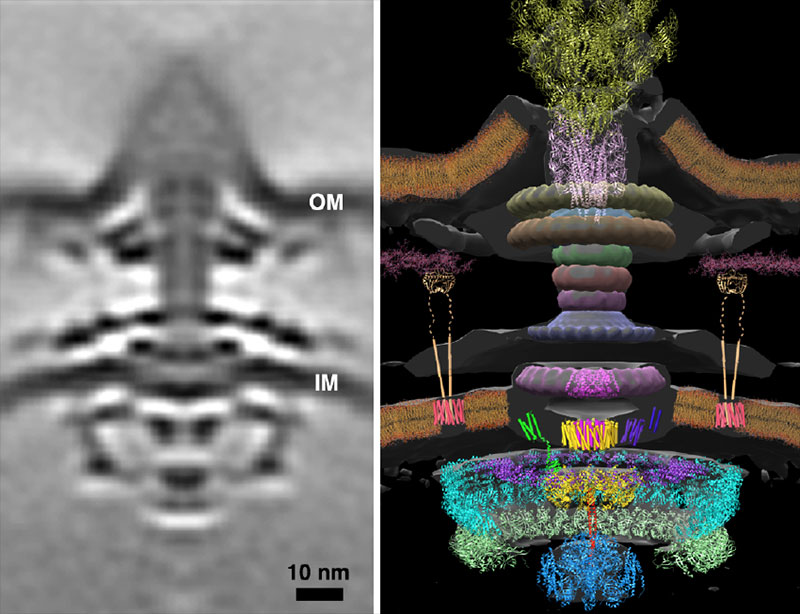
Subtomogram average (left) and molecular model (right) of a bacterial flagellar motor. CryoET will advance our understanding of such molecular machines.
The Schmidt Academy for Software Engineering collaborated with the Jensen group on the development of open-source software with two main goals in mind: 1) increased automation of the ECT data processing pipeline and 2) more rigorous evaluation of existing (and often competing) algorithms/tools in the field through standardized comparison. To this end, two distinct bodies of work were produced and shall be discussed below.
Aritatomi
In a collaboration with the Frangakis Group at the Buchmann Institute for Molecular Life Sciences - Goethe University Frankfurt, the Schmidt Scholar helped to professionalize a novel CET processing package called Artiatomi so that it could be distributed to researchers outside the developing lab (including the Jensen group) and thus extensively evaluated. Boasting intriguing new algorithms and GPU-accelerated computational components, Artiatomi presented a clear candidate for applying software best practices to help bring it to the greater community. The Schmidt Scholar was able to take this research software that could not be compiled outside the originating lab's computing environment to a modern, easily-distributable package. Specifically, the Scholar refactored the package into a well-structured CMake project and set-up public deployment infrastructure for vital internal documentation and Matlab driver scripts that had not been made available before. Additionally, the Scholar made available an easy-install version of the Artiatomi package leveraging Docker containerization and user-friendly wrapper scripts to abstract away the complexities of Docker setup for the average biologist.
The cleaned-up, official Artiatomi package is made available at: https://github.com/uermel/Artiatomi
The Dockerized version of Artiatomi (artiatomi-tools) packaged with various helper scripts is available at: https://github.com/kmshin1397/artiatomi-tools
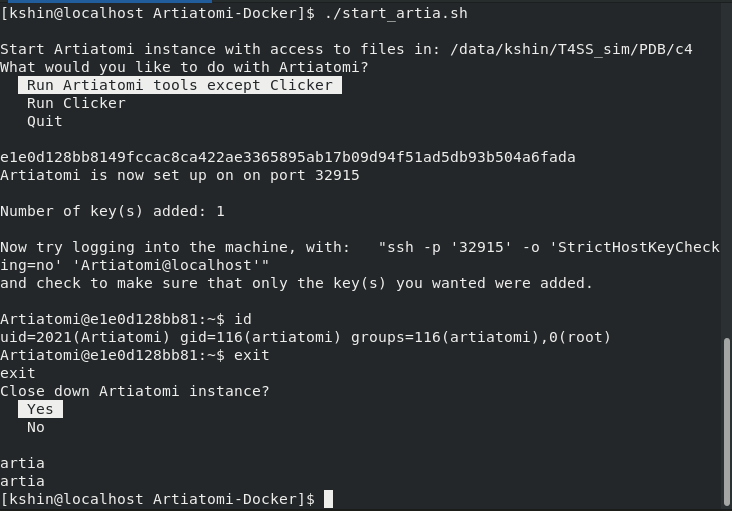
A screenshot of artiatomi-tools spawning a Dockerized Artiatomi instance and automatically setting up an SSH connection to it
ETSimulations
The ETSimulations project was developed to address the goals of automation and benchmarking with a focus on the context of simulated CET data. ETSimulations is both a simulation and processing platform for cryoelectron tomography. Its simulation module provides easy set-up for simulation of large CET data sets by orchestrating many parallel instances of external software which handle the various scientific computation steps involved in the generation of simulated raw data. ETSimulations also provides a processing module to help automate much of the processing of the simulated raw data, allowing the user to choose from a collection the most interesting CET processing software packages currently available. Given the extra known information afforded by simulation data, much more of the processing pipeline can be automated to a high-degree of competence when compared to working with real data - resulting in much higher relative throughput. This and the fact that we know the known structures that went into the simulations means that ETSimulations provides a powerful tool for comparing and contrasting the variety of processing tools available in the field. Additionally, the automatic processing modules in ETSimulations has been adapted and made available for use with real data projects as well where possible to improve general group-wide productivity.
The ETSimulations package is made available at: https://github.com/kmshin1397/ETSimulations
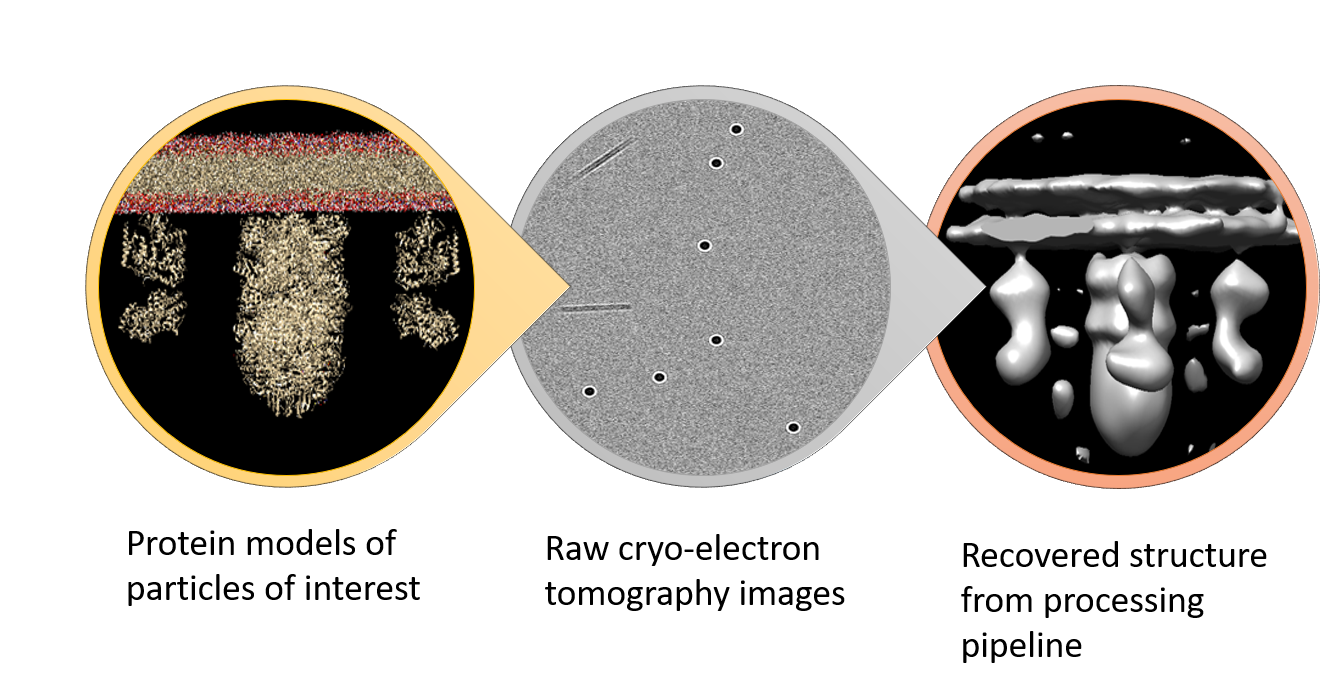
Example images from the ETSimulations workflow