Projects
flyfish and nymph: Spatial Genomics Packages
SASE: Konnor von Emster (Division of Biology and Biological Engineering), SASE GRA Fellow
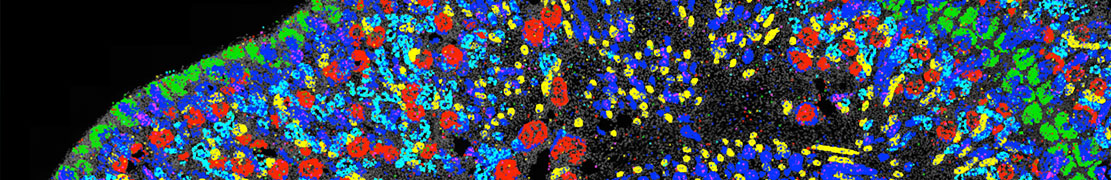
Spatial genomics is a field focused on producing molecular maps of gene activity within intact tissues. Such maps allow scientists to study how cells are organized and how they influence one another, enabling direct investigation of organ development, disease progression, and cell–cell communication. At Caltech, the Cai lab is a leader in improving spatial genomics technologies such as seqFISH that generate these rich datasets. Despite major advances in such experiments, the software needed to design, run, and analyze these experiments remains fragmented, difficult to maintain, and challenging for labs to adopt.
To address this gap, the Schmidt Academy for Software Engineering (SASE) is supporting the development of flyfish and nymph, two integrated software tools designed to make spatial genomics more accessible, reliable, and scalable. Flyfish provides a modular toolkit for experiment design, execution, and analysis, allowing researchers to assemble customized workflows or use individual components independently. Nymph builds on these outputs to interpret the resulting molecular maps by identifying structural patterns in tissues, modeling developmental changes, and highlighting genes involved in key transitions. Together, these tools will establish a coherent, end-to-end platform for spatial genomics that can be used broadly across the biological research community.
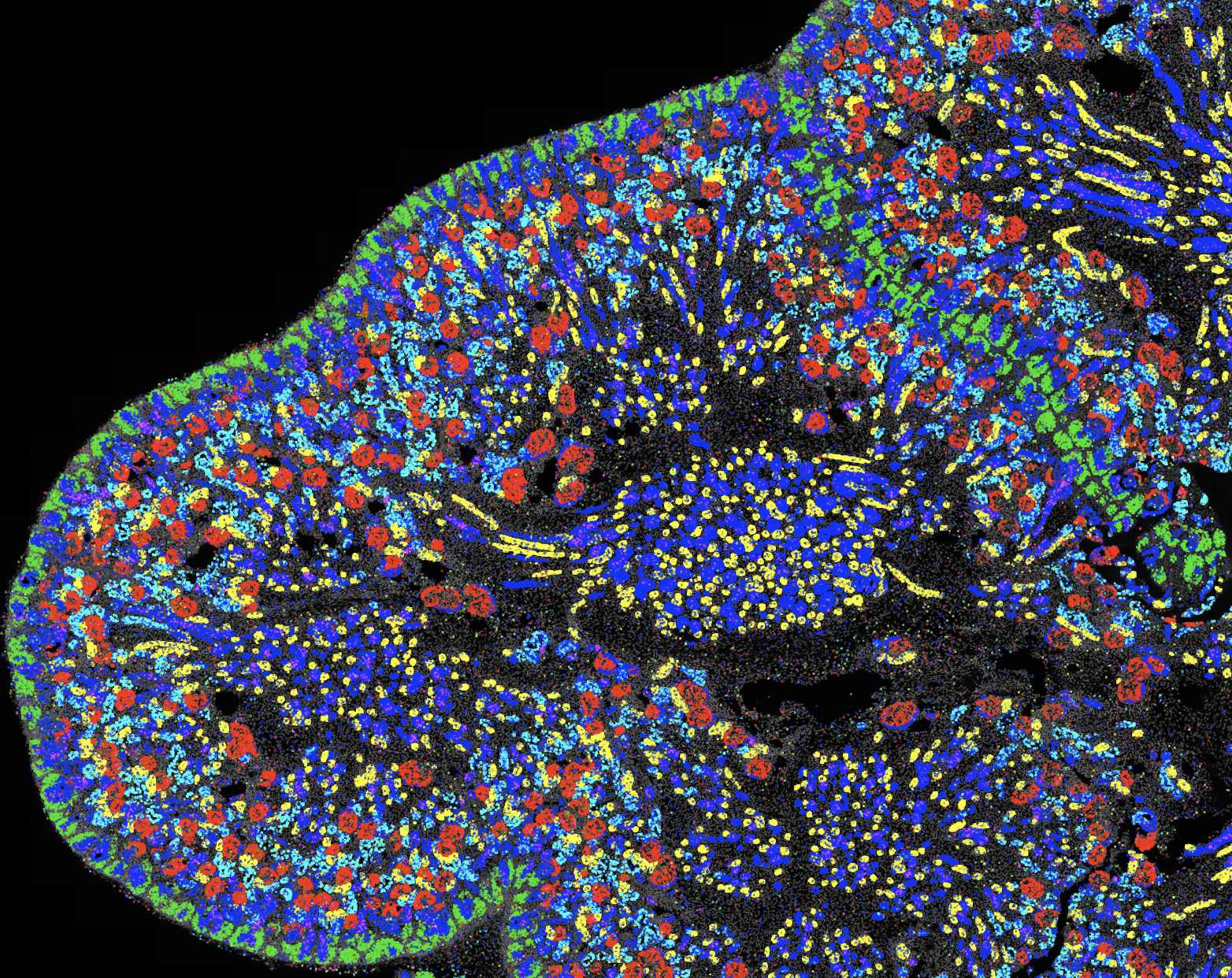
Developing software that can support the multi-terrabyte scale and complexity of modern spatial genomics data, operate consistently across computing environments, and remain maintainable for long-term community use presents substantial engineering challenges. SASE mentorship will focus on improving testing, standardizing containerization, clarifying module boundaries, and expanding documentation to strengthen reliability and sustainability. User-friendly interfaces and tutorials will further ensure that the tools can be adopted by groups without extensive computational backgrounds. By the end of the fellowship, these resources will provide a scalable, community-ready toolkit for spatial genomics and support more systematic, quantitative analysis of large-scale spatial data.