
Projects
DiazoDB: An Interactive Website to Explore Nitrogenase Sequence Diversity
SASE: Zahra Shivji (Division of Engineering and Applied Science), SASE GRA Fellow
Nitrogenase, the sole enzyme responsible for biological nitrogen fixation, is actively being explored for sustainable ammonia production via bioelectrocatalysis or as a biological crop amendment. However, engineering nitrogenase for commercial use presents additional challenges: Its low thermostability, tight regulatory control, and bulky gene set (>50 nitrogen fixation genes in the model organism Azotobacter vinelandii) make it challenging to deploy outside native hosts. To overcome these challenges, extant nitrogenase diversity was systematically characterized to understand the enzyme’s regulatory mechanisms, evolutionary history, and environmental adaptations (such as thermotolerance and resilience against unfavorable redox conditions). This effort represents the most extensive annotation of nitrogenase to date, identifying six times more variants (~6,000 nitrogenase operons, nifHDKEN) compared to conventional approaches. Zahra is collaborating with SASE to develop two interactive websites. To advance protein engineering goals, these findings will be made publicly available on an interpretable and interactive website, DiazoDB, for future phylogenetic and functional analysis of these enzymes.
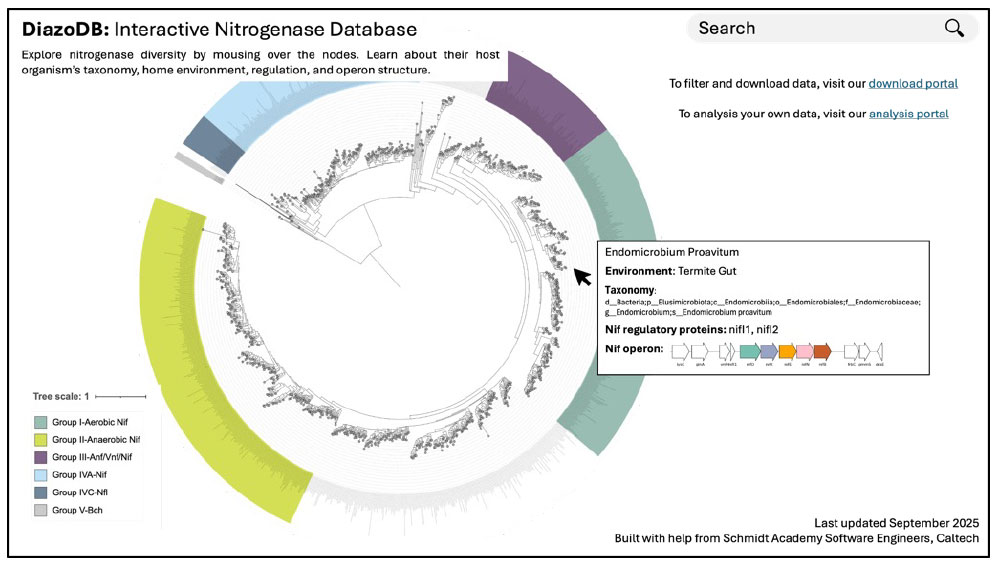
DiazoDB mockup: The main webpage will host an interactive nitrogenase phylogenetic tree. Mousing over the nodes will provide information about the host organism’s taxonomy, environment, nitrogenase regulation, and operon structure.
DiazoDB (named for diazotrophs, organisms that fix nitrogen) will be a website to explore the diversity and evolutionary history of over 6,000 nitrogenase sequences. The main webpage will host an interactive nitrogenase phylogenetic tree. Mousing over the tree’s nodes will provide detailed metadata on each sequence, including taxonomy, environment of origin, regulatory context, and gene diagrams for the nif operon. By providing the ability to easily browse known nitrogenase sequence and organismal diversity, DiazoDB will become a foundational tool for enzyme engineers, evolutionary biologists, and microbial ecologists. Currently, no web-based interactive trees exist for proteins, despite their significance to Earth’s biogeochemical processes and human health; DiazoDB will serve as a model tool for protein data accessibility. Collaboration with the Schmidt software engineers will ensure that DiazoDB is not only publishable but also a durable, open-access resource for protein engineers, microbial ecologists, and evolutionary biologists to advance their research.