
Projects
Deep Cell - CellSAM
Deep learning-enabled software for single-cell analysis
PI: David Van Valen (Division of Biology and Biological Engineering)
SASE: Raffey Iqbal, Scholar
Single-cell imaging captures the temporal and spatial variations that underlie life, from the scale of single molecules to entire organisms. Advances in multiplexed imaging have enabled the simultaneous measurement of multiple proteins in situ, offering unprecedented visualization of cell types and states within tissues. The Van Valen Lab's software suite, DeepCell, utilizes a variety of deep learning models trained to perform single-cell analysis tasks at levels equal to or superior to those of humans, ranging from segmenting cells in pathology images to identifying individual cell phenotypes. These models render difficult analyses routine and empower new, previously impossible experiments across the life sciences field. As part of this project, the Schmidt Academy Scholar contributed to several key deep learning models of the DeepCell suite.
First, they modernized a legacy cell segmentation package, Mesmer. Mesmer, having been trained on TissueNet (with over a million hand-labeled cells), was one of the first models that could segment whole cells in diverse tissue images with human-level accuracy, which made downstream tasks, like measuring protein localization and quantifying tumor–immune architecture, reliable and repeatable across labs and imaging platforms. The scholar’s work on re-implementing it from scratch in PyTorch provides long-term usability and compatibility, and ensures that the valuable research software built on top of Mesmer continues to work seamlessly.
Next, the Scholar worked on CellSAM, DeepCell’s current state-of-the-art cell segmentation model. CellSAM, built by pairing SAM with an object detector, improves upon Mesmer and enables robust, promptable segmentation that scales, which is essential for analyzing whole slide images. The Scholar worked on extending CellSAM with the ability to split images into smaller sections, segment them individually, and seamlessly stitch the results together. Doing so made it more memory efficient and allowed the segmentation of whole-slide images that would otherwise be computationally prohibitive.
Finally, the Scholar worked on training DeepCellTypes. A cell phenotyping package that uses a language-informed vision model to learn what each antibody marker “means” and so addresses the key bottleneck of consistent phenotyping across experiments that use different antibody panels. On this sub-project, the Scholar developed data processing pipelines, optimized training code, and designed and ran experiments to ensure generalizability and robustness of the model.
Together, these changes improved software sustainability, allowed analysis of much larger samples, and completed the development of a cell phenotyping model that can allow cell and tissue analysis for diverse antibody panels and imaging techniques.
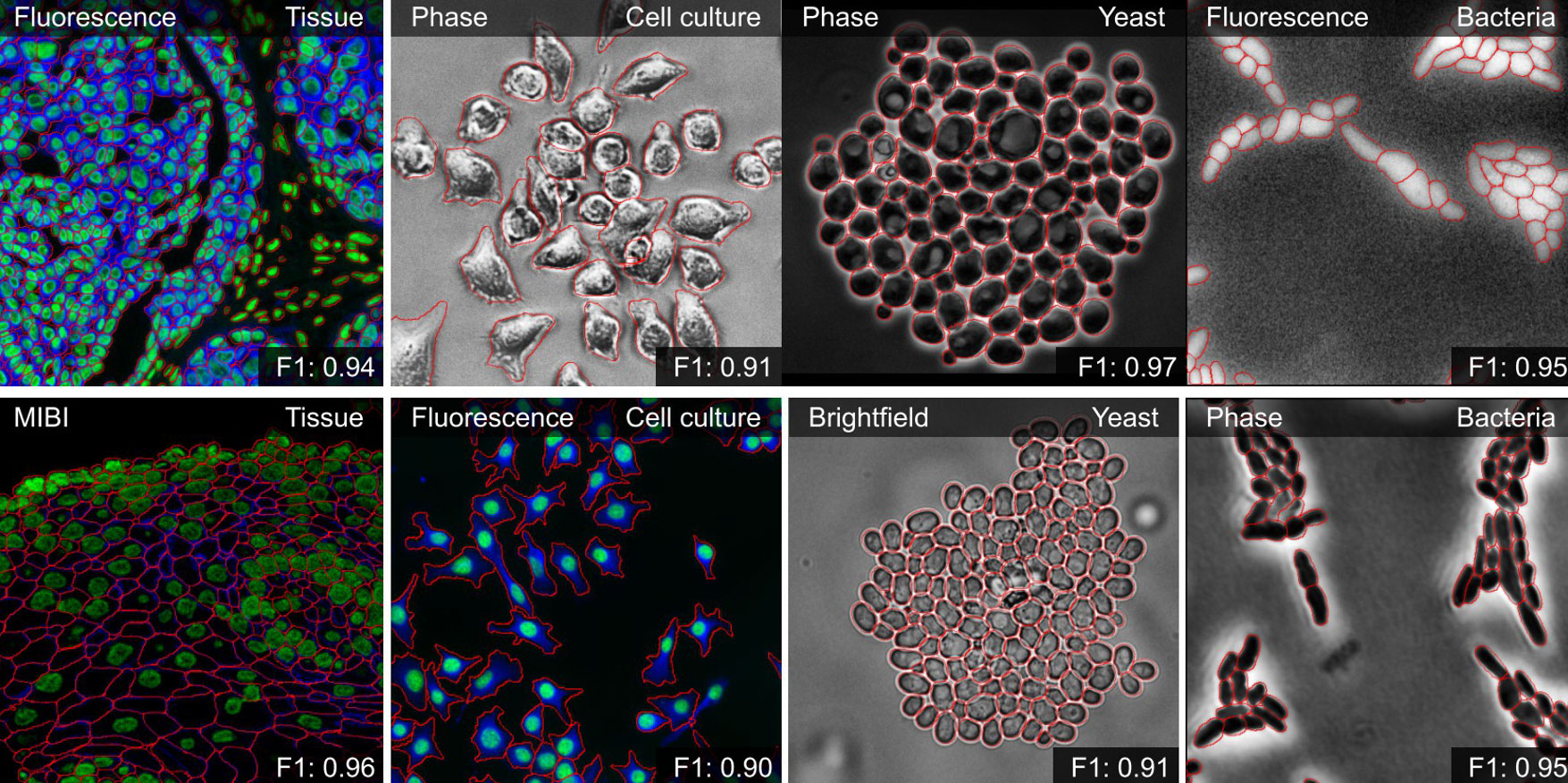
Fig. 1 - Qualitative results of CellSAM segmentations for different data and imaging modalities. Predicted segmentations are outlined in red.
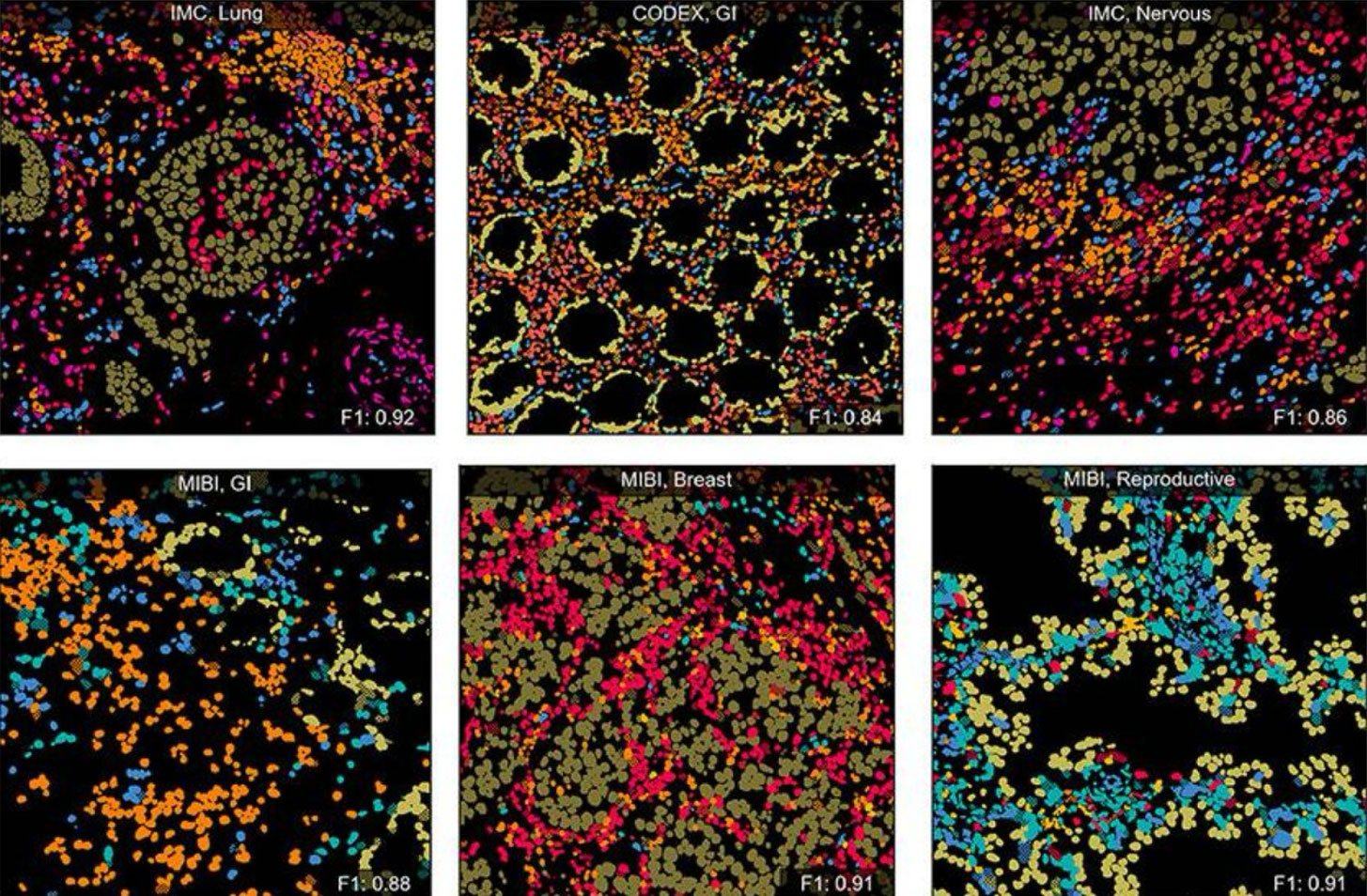
Fig. 2 - Qualitative results of DeepCellTypes in example Field of Views (FOVs) with cell types colorized